About
I'm interested in science, evolution, virology, and the development of bioinformatics tools and workflows. I have a degree in Biotechnology but my expertise is in Bioinformatics. I hold a Ph.D. in Sciences from Fundação Oswaldo Cruz (FioCruz), I'm a bioinformatician volunteer at Rede Genômica FioCruz (RGF - Fiocruz Genomic Network) and work as Bioinformatician Analyst at Genesis Genomics, in Brazil.

Bioinformatician
My work on Genesis Genomics involves the development and application of bioinformatics workflows to analyze human genomics (germline and somatic variants) and code development to automatize tasks on AWS. To do it, I mainly develop with Workflow Development Language (WDL), Amazon Web Services (AWS), Docker, Git, Shell Script and Python in agile teams following the Scrum methodology.
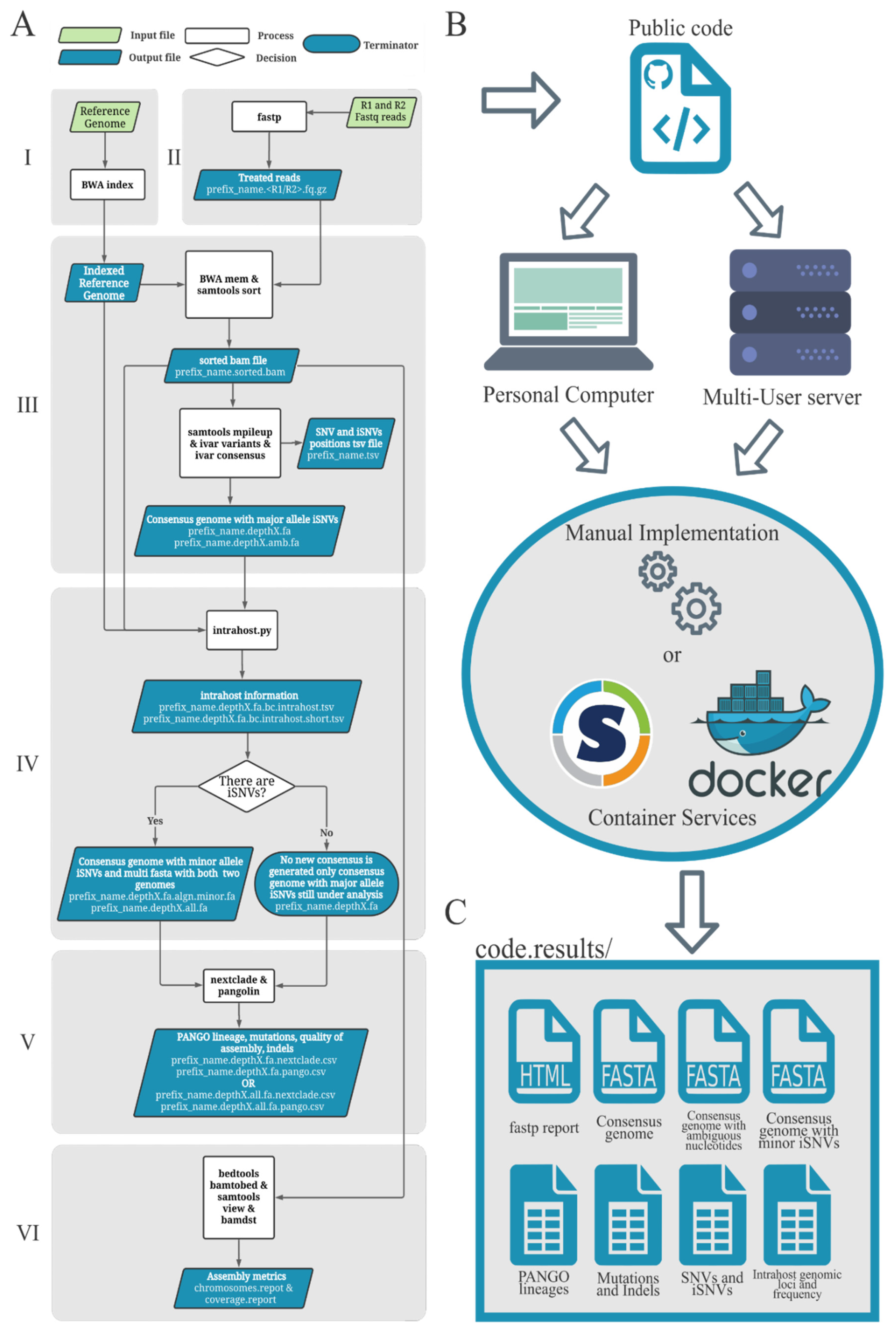
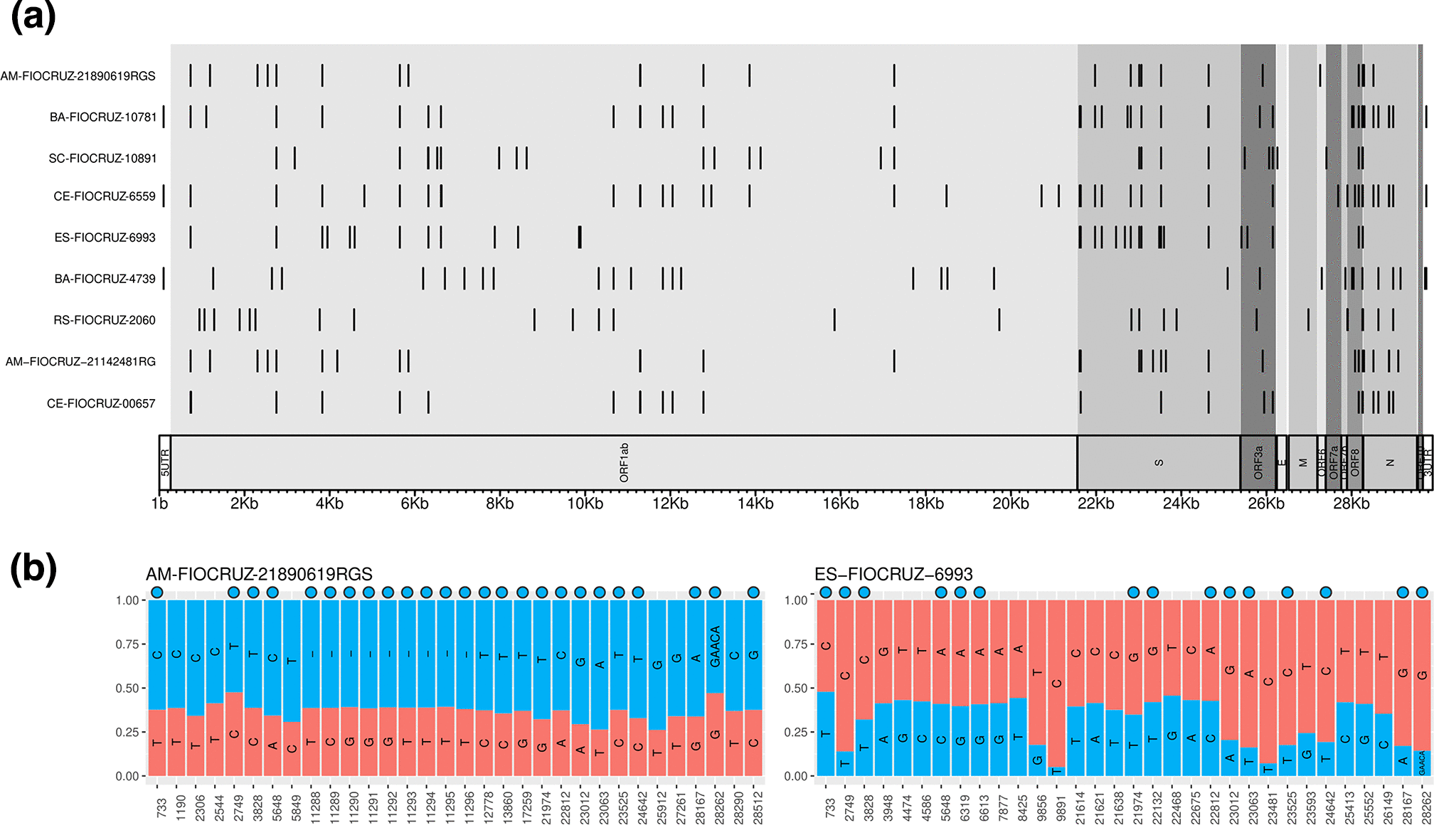
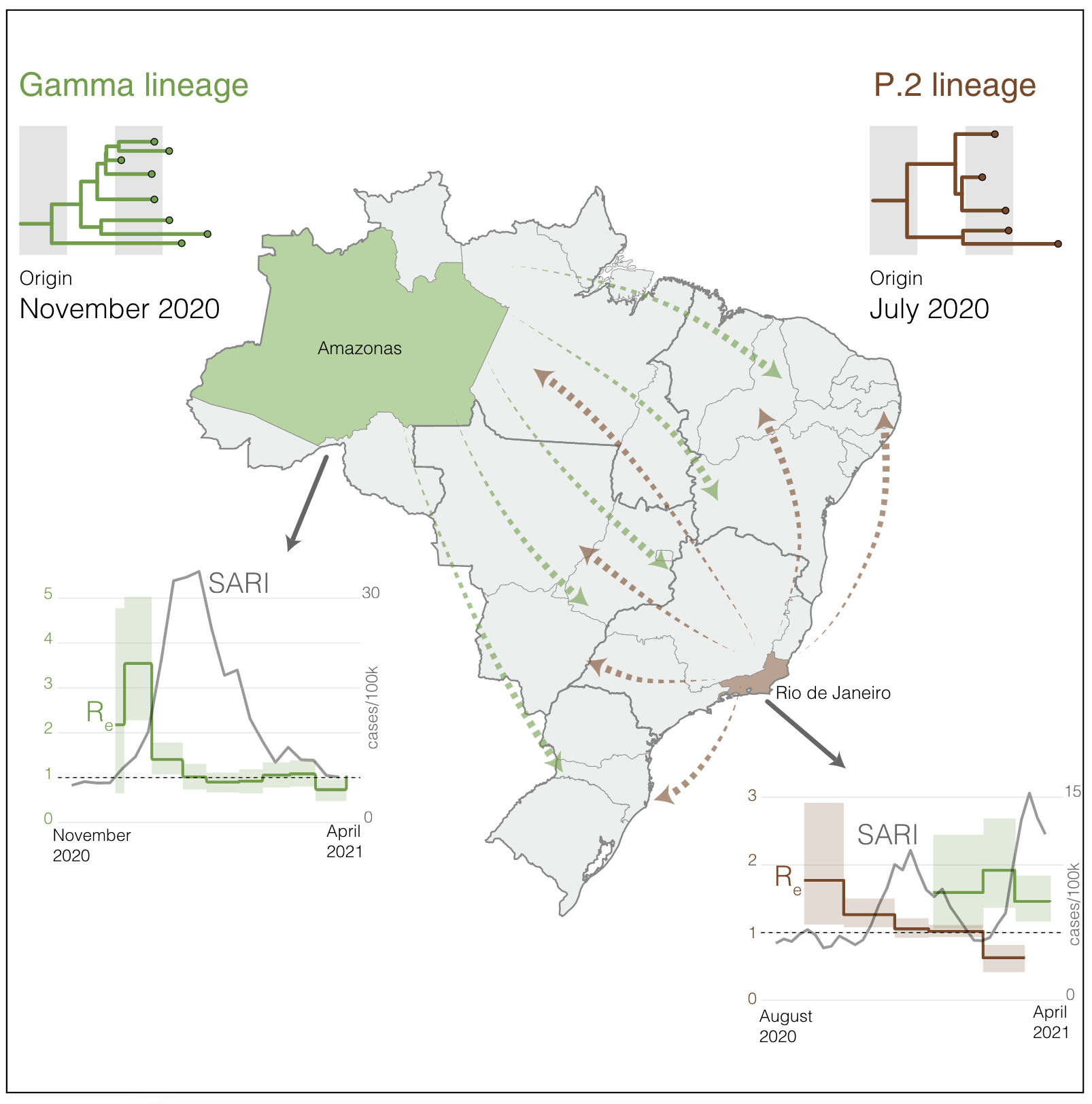
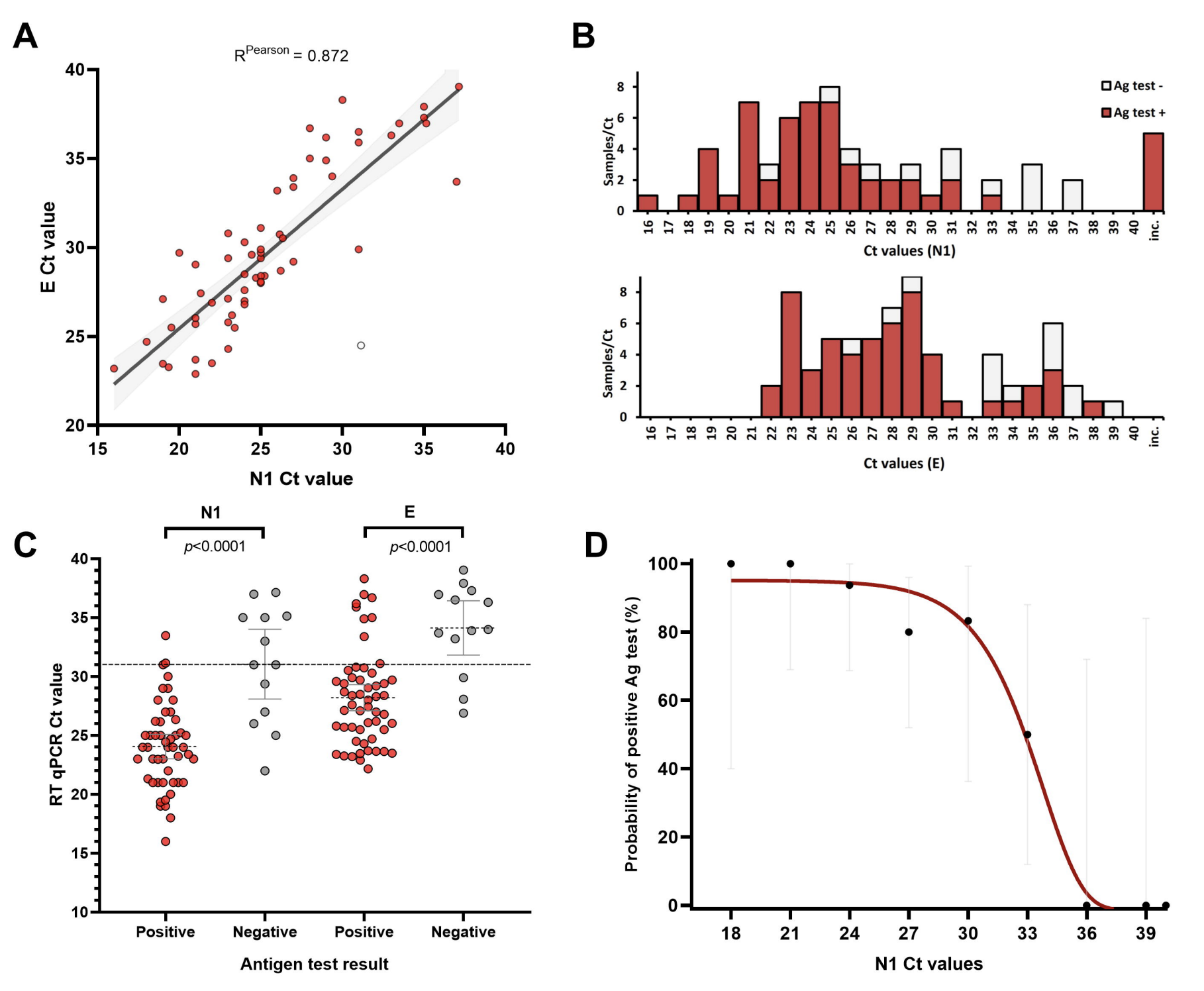
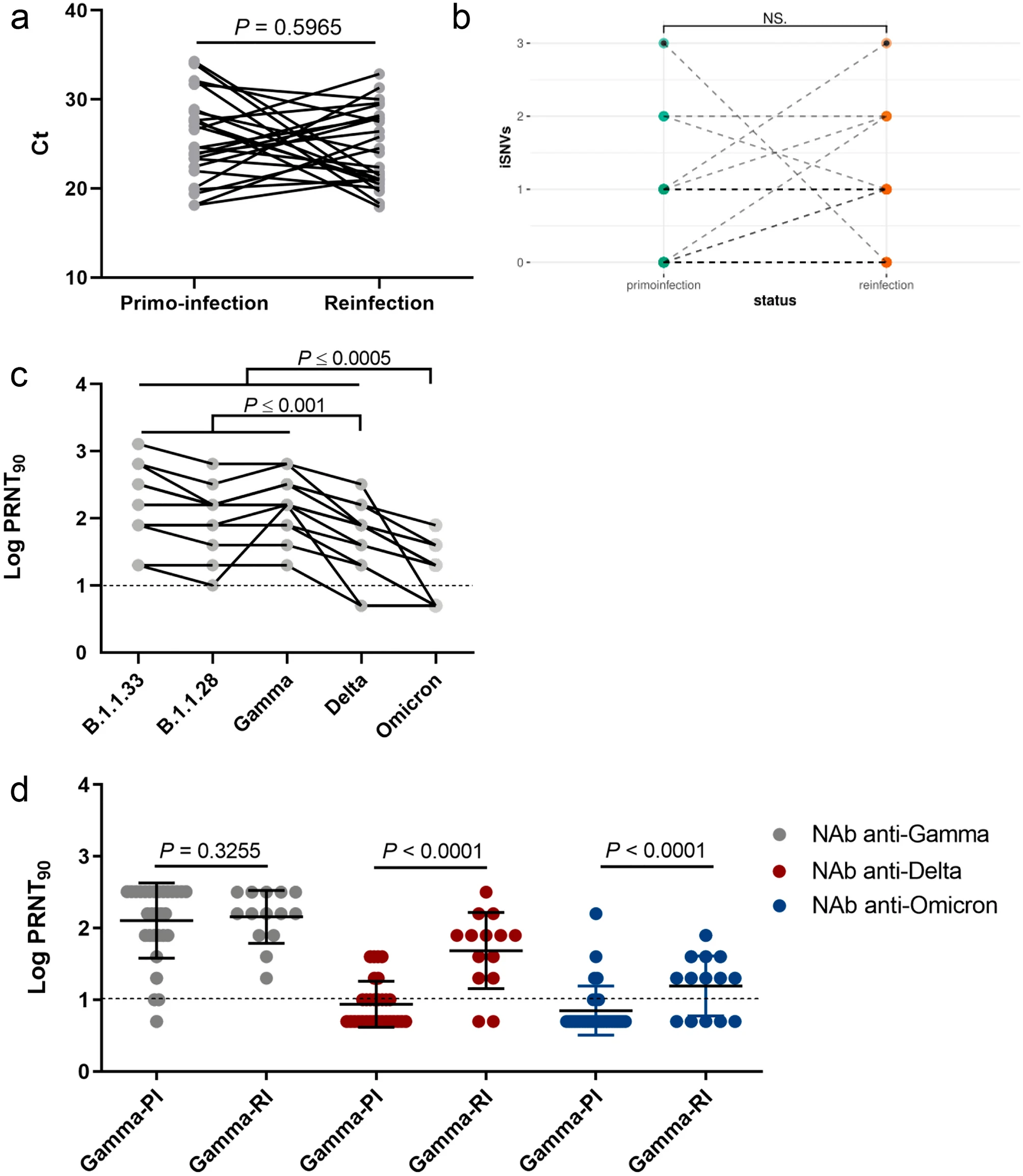
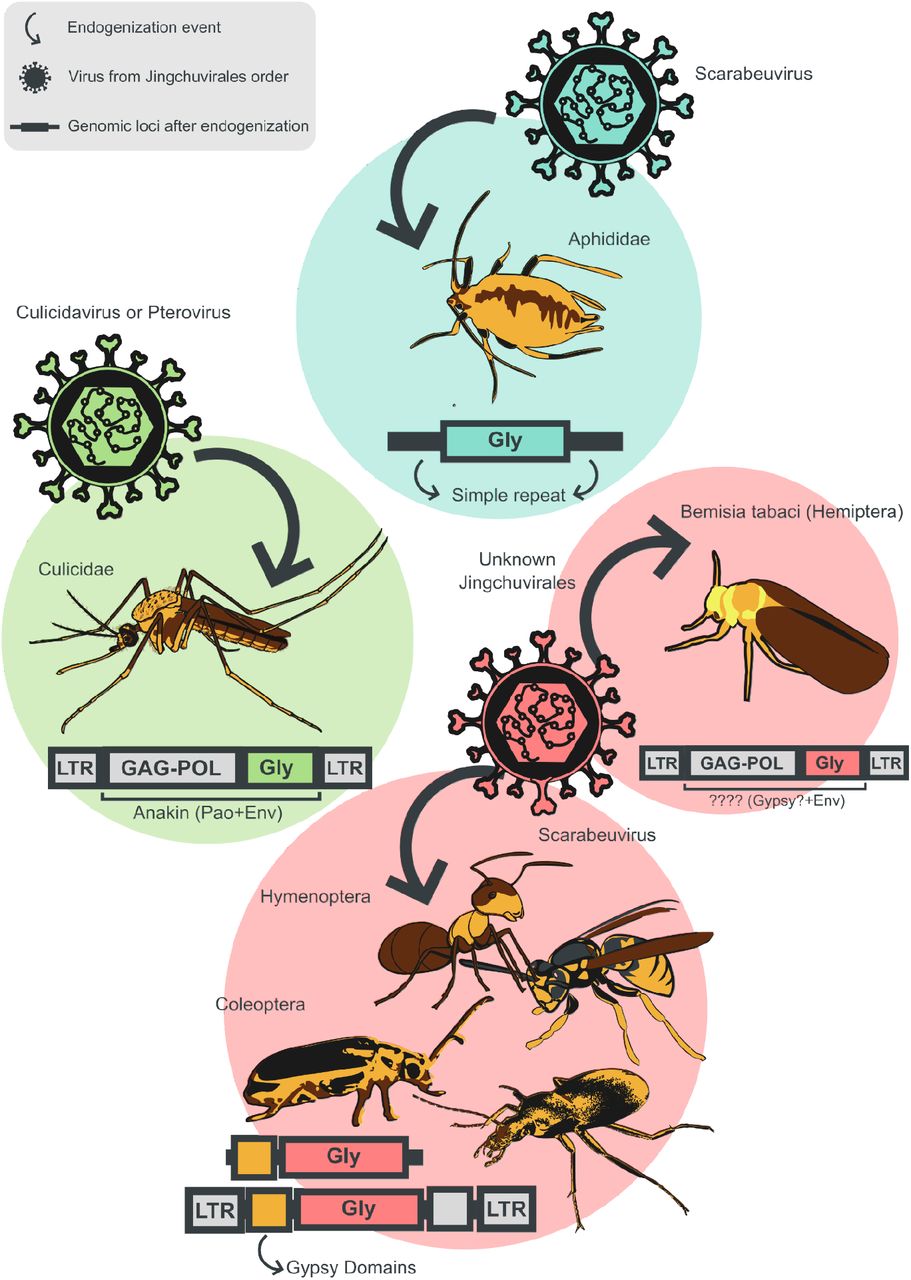
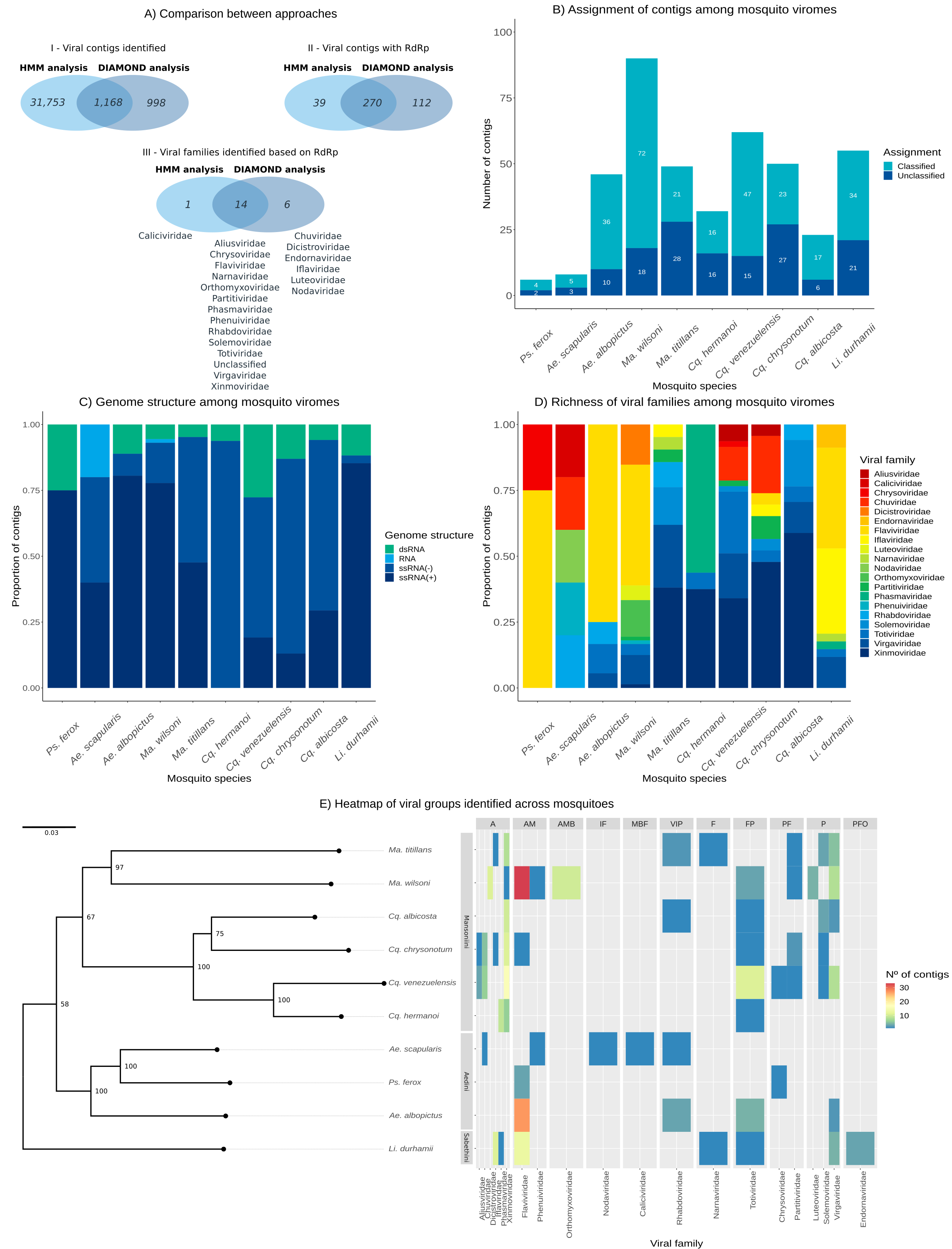
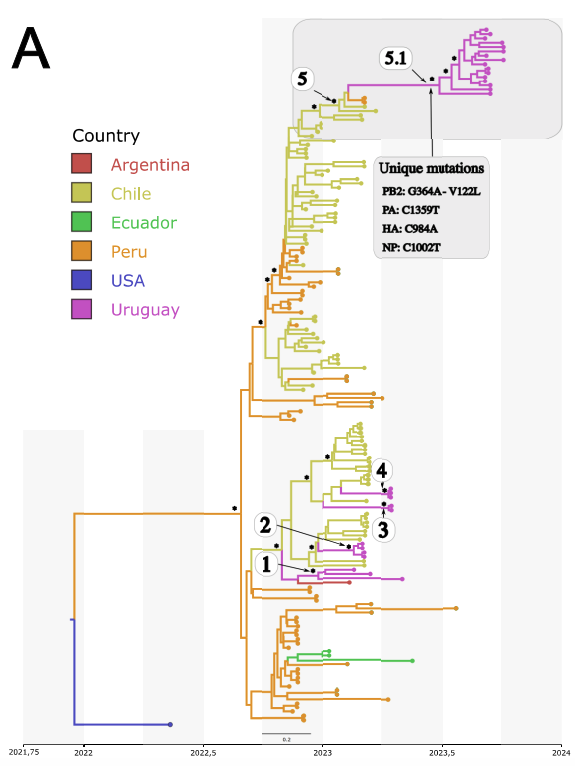
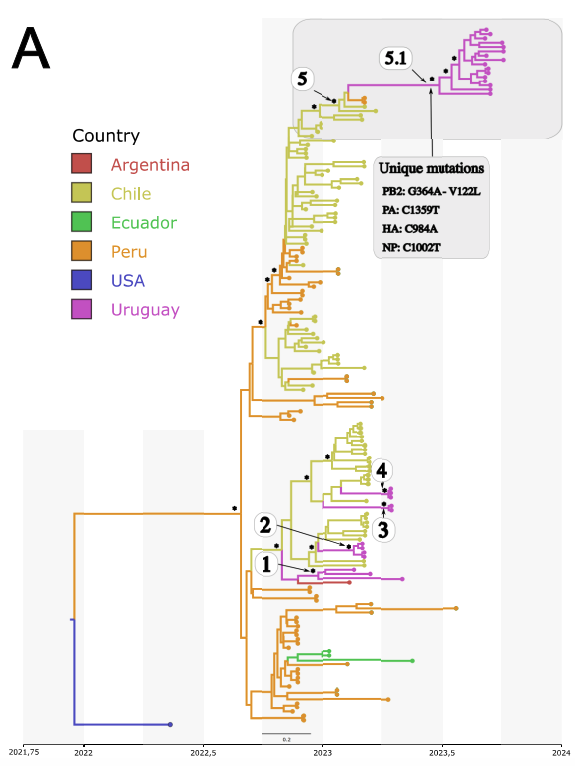
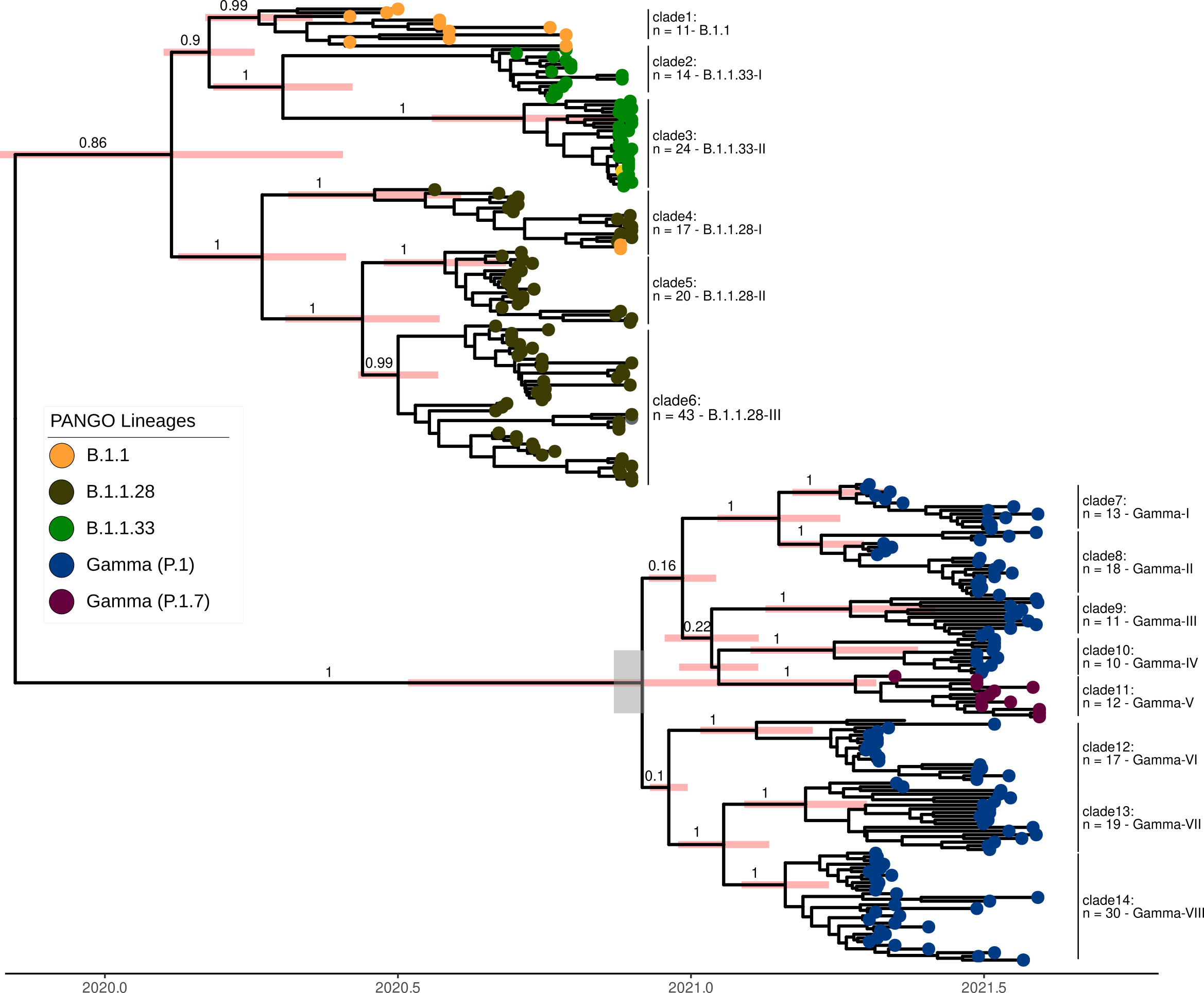
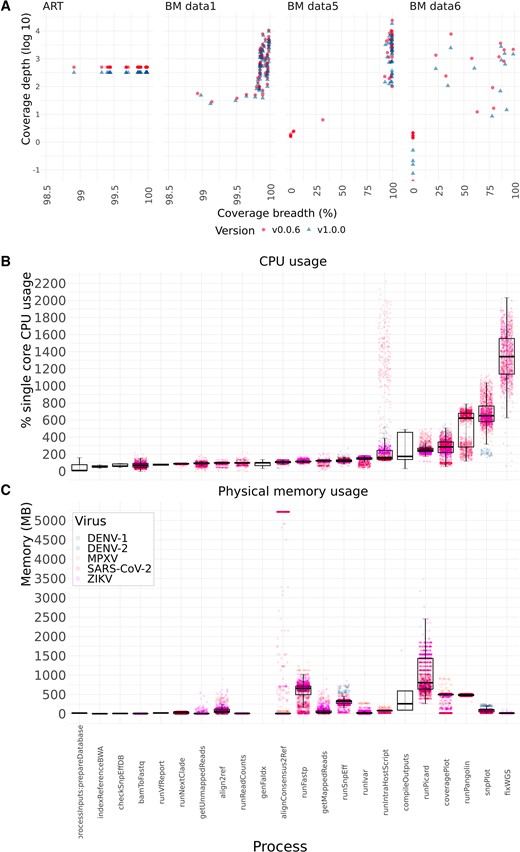
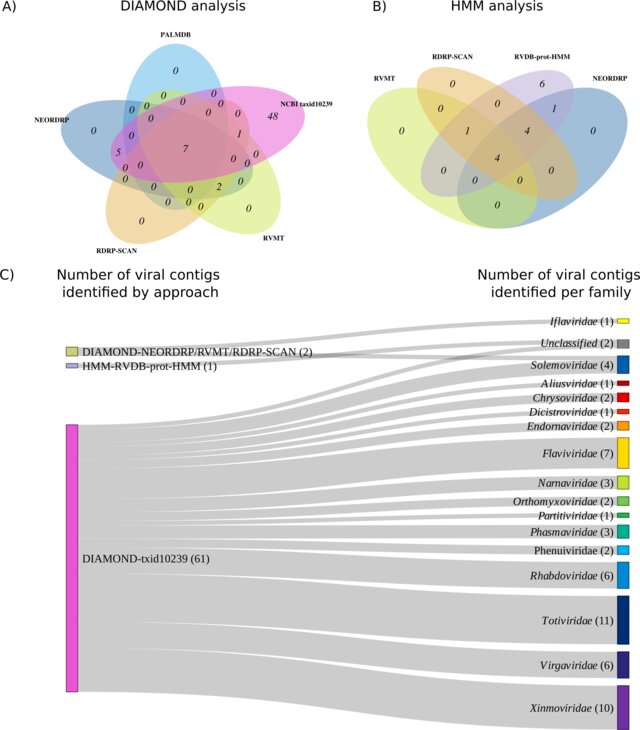
During my Ph.D., I worked with genomic surveillance of SARS-CoV-2. I built the ViralFlow - with other members of RGF - a Nextflow workflow that performs a set of viral genomic analyses from sequencing raw data. Moreover, I published key studies to understand several aspects of SARS-CoV-2 evolution during the pandemic in Brazil as well as I collaborated with several studies mainly on the biology of mosquito vectors of arbovirus and paleovirology.
To perform my Ph.D. research I worked with Multi-user Linux servers, PBS queue system, Docker, Singularity, Nextflow, Python, R and Shell Script.
In my free time, I like creating and sharing content related to bioinformatics on Linkedin, reading books, drawing and last but not least, I like having a good time with my family and friends.
Resume
A brief story of my journey.
Sumary
Filipe Dezordi
Bioinformatician Analyst with 4+ years of experience, developing and applyng bioinformatics tools and workflow to study viral and human genomics.
- Curitiba, Paraná, Brazil.
- filipe.zimmer@fiocruz.br
Education
Ph.D. in Biosciences and Biotechnology on Health
2020 - 2024
Fundação Oswaldo Cruz (FioCruz) - IAM, PE
My Ph.D. project research was about SARS-CoV-2 genomics and evolution. During this period, I did my research on the Bioinformatics Core of FioCruz and became a member of Rede Genomica FioCruz (FioCruz Genomic Network). I worked on a multi-user cluster with PBS system, I improved my skills in Python and Shell Script programming, as well as learned R, Docker, and Singularity. I developed - with the RGS staff - the ViralFlow, the workflow that was used to analyze more than 38k genomes in Brazil during the SARS-CoV-2 pandemic.
Master Degree in Biosciences and Biotechnology on Health
2018 - 2020
Fundação Oswaldo Cruz (FioCruz) - IAM, PE
My master's degree research was about Endogenous Viruses Elements in mosquito genomes. During this period, I did my research on the Entomology Department and Bioinformatics Core of FioCruz. I worked on multi-user Linux servers and started programming in Python and Shell Script.
Bachelor of Biotechnology
2013 - 2017
Universidade Federal do Pampa, São Gabriel, RS
During this period I started in the research field working for 3 years as a volunteer at Proteomics Applied Lab, where I studied Transposable Elements in a parasitoid wasp draft genome, using Galaxy and GUI software.
Professional Experience
Full Bioinformatician Analyst
2024 - Present
Genesis Genomics, Brazil, Remote
My current position. In April 2024, the departament in which I worked at the Hospital Israelita Albert Einstein went through a merger process, creating the Genesis Genomics, to which I was transferred to continue working in the same position
- Automations and Integrations staff.
- Daily development of codes using Python.
- Development and update of python CLI tools / packages.
- Development of serverless systems using AWS CDK in python.
- CI/CD with github actions and AWS code pipelines.
Full Bioinformatician Analyst
2022 - 2024
Hospital Israelita Albert Einstein (HIAE - Varsomics), Recife, PE
- Routines of bioinformatics analysis applied to precision medicine.
- Daily development of codes using Python and WDL.
- Worked on codes automatize tasks on AWS, and to include new features on Human genomics workflows.
- Integrate the BioDevOps team, building AWS infraestructure using AWS CDK (IaC).
- In september 2023 change to full remote position.
- In April 2024 transferred to Genesis Genomics.
Volunteer Bioinformatician
2020 - Present
Rede Genomica FioCruz (RGF), Recife, PE
- Workflow development and maintanance.
- SARS-CoV-2 genomic data analysis, foccused on intrahost variants and large phylogenies.
- Rmarkdown reports to governamental institutions.
Publications
My papers as main or collaborator author published in Scientific Journals.
- All
- 2018
- 2020
- 2021
- 2022
- 2023
- 2024
Codding
Repositories for tools, scripts or websites that I`ve developed and are publicly available.